In this post I’ll go through the simple implementation of LRA (Logistic Regression Algorithm) to outline the Phylanx architecture and also demonstrate how one might go about writing their own programs in Phylanx. The complete version of the code discussed in this post can be found in the project’s GitHub repository under examples/algorithms directory and the corresponding dataset can be found here.
Phylanx Architecture
The core of Phylanx has been implemented in HPX (a C++ Standard Library for Concurrency and Parallelism), and has three components (figure 1):
- PhySL– a minimal language developed for testing purposes (the syntax is subject to change).
- Phylanx AST- constructed by the Phylanx (PhySL) parser and will be used for analyses and optimizations.
- Execution Tree- compiled from the AST and will run asynchronously for maximum performance.
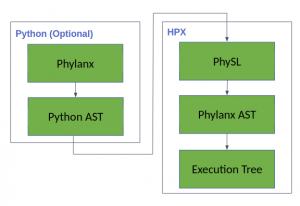
Figure 1. Phylanx Components
We recognize the fact that Phylanx will mostly be used by domain experts working on massive datasets. Therefore, to lift the burden of learning a new language, we also provide a Python interface (frontend) to Phylanx. The Python frontend enables programmers to write their code once, in a familiar language, and run it on any platform (whether a PC or large cluster) without changing the source code. At the moment this feature is available through the use of Python decorators. Any valid Python code with Phylanx constructs can be wrapped into a function which in turn is decorated with *@phyfun* function (the name is subject to change). @phyfun will automatically generate the equivalent PhySL string, and subsequently compile, and evaluate it.
LRA Example
In this example we will demonstrate how to write an LRA with the Python frontend. We will use a breast cancer diagnostic dataset as an input to our algorithm. This dataset contains 569 rows (1 for each patient) with 30 columns containing the features and one last row with the diagnosis.
As mentioned above, any code snippet to be translated into PhySL must first be wrapped in a function. Here, we call the function lra and decorate it with @phyfun.
1 2 3 4 5 6 7 8 9 10 11 12 | import phylanx from phylanx.util import * @phyfun def lra(file_name, xlo1, xhi1, ylo1, yhi1, xlo2, xhi2, ylo2, yhi2, alpha, iterations, enable_output): # <code to be transformed into PhySL> return weights |
Now we can write our program as the body of the lra function and Phylanx will automatically translate the code into PhySL.
As the first step we need to read data which is stored in CSV format. Phylanx provides a primitive (function) read_csv_file to read this type of file. We read the file and store the feature values in matrix x and diagnoses in vector y:
1 2 3 | data = file_read_csv(file_name) x = data[xlo1:xhi1,ylo1:yhi1] y = data[xlo2:xhi2,ylo2:yhi2] |
Next we’ll initialize the prediction, gradient and error vectors to zero using the constant function:
1 2 3 | pred = constant(0.0, shape(x, 0)) // prediction error = constant(0.0, shape(x, 0)) gradient = constant(0.0, shape(x, 1)) |
We also store the transpose of the x as we’ll use it many times later within the while loop:
1 | transx = transpose(x) |
Once everything is initialized and loaded, we can start the iterations to find the (optimal) weight function. Within the loop, we calculate the prediction (using Phylanx functions exp and dot), error, gradient (using dot) and weight. One should note that division, summation, negation are all also Phylanx primitives.
1 2 3 4 5 6 | while step < iterations: pred = 1.0 / (1.0 + exp(-dot(x, weights))) error = pred - y gradient = dot(transx, error) weights = weights - (alpha * gradient) step += 1 |
Now, when the lra function is called, Phylanx will automatically generate the PhySL and evaluate it. We can print the result using phy_print function.
1 2 | res = lra("breast_cancer.csv", 0,569,0,30, 0,569,30,31, 1e-5,750,0) phy_print(res) |
We have also done some preliminary performance analysis of our toolchain. We found that the performance achieved by Phylanx is comparable to that of numpy:
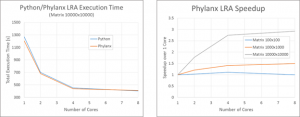
We are confident that as we continue to expand and improve Phylanx we will be able to pare down our execution time and improve upon the scalability we currently observe.